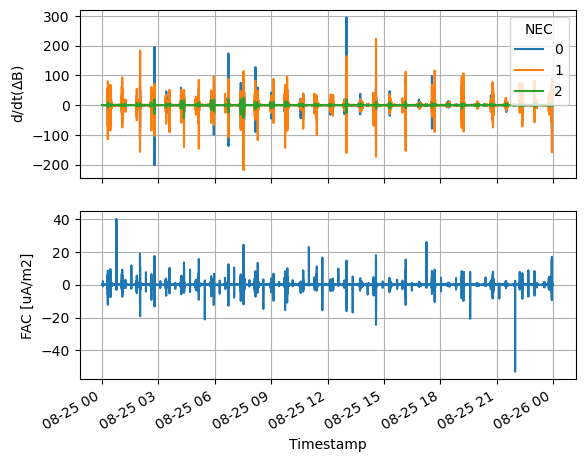
Demo FAC on-demand quicklook#
Using SwarmPAL implementation in development. See https://swarmpal.readthedocs.io/en/latest/guides/fac/intro_fac.html
This example uses HAPI instead of the VirES API
import datetime as dt
from hapiclient import hapi
import numpy as np
from swarmpal.io import create_paldata, PalDataItem
from swarmpal.toolboxes import fac
SERVER = "https://vires.services/hapi"
DATASET = "SW_OPER_MAGA_LR_1B"
Find latest available time#
meta = hapi(SERVER, DATASET)
latest_available = dt.datetime.fromisoformat(meta["stopDate"])
latest_available
datetime.datetime(2024, 8, 25, 23, 59, 59, tzinfo=datetime.timezone.utc)
Fetch latest available day#
start = latest_available - dt.timedelta(days=1)
stop = latest_available
data = create_paldata(
PalDataItem.from_hapi(
server=SERVER,
dataset=DATASET,
parameters="Latitude,Longitude,Radius,B_NEC,B_NEC_Model",
start=start.strftime("%Y-%m-%dT%H:%M:%S"),
stop=stop.strftime("%Y-%m-%dT%H:%M:%S"),
)
)
# Temporary fix to data labelling
data = data.rename_dims(
{"B_NEC_dim1": "NEC", "B_NEC_Model_dim1": "NEC"}
)
print(data)
/opt/conda/envs/swarmpal-runner/lib/python3.11/site-packages/hapiclient/hapitime.py:287: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
Time = pandas.to_datetime(Time, infer_datetime_format=True).tz_convert(tzinfo).to_pydatetime()
DataTree('paldata', parent=None)
└── DataTree('SW_OPER_MAGA_LR_1B')
Dimensions: (Timestamp: 86400, NEC: 3)
Coordinates:
* Timestamp (Timestamp) datetime64[ns] 2024-08-24T23:59:59 ... 2024-08-2...
Dimensions without coordinates: NEC
Data variables:
Latitude (Timestamp) float64 -3.933 -3.87 -3.806 ... 65.26 65.2 65.13
Longitude (Timestamp) float64 -35.61 -35.61 -35.61 ... 137.5 137.5 137.5
Radius (Timestamp) float64 6.847e+06 6.847e+06 ... 6.834e+06 6.834e+06
B_NEC (Timestamp, NEC) float64 1.865e+04 -6.146e+03 ... 4.655e+04
B_NEC_Model (Timestamp, NEC) float64 1.865e+04 -6.144e+03 ... 4.655e+04
Attributes:
PAL_meta: {"analysis_window": ["2024-08-24T23:59:59", "2024-08-25T23:59:...
Apply FAC algorithm#
process = fac.processes.FAC_singlesat(
config={
"dataset": DATASET,
"model_varname": "B_NEC_Model",
"measurement_varname": "B_NEC",
},
)
data.swarmpal.apply(process)
print(data)
DataTree('paldata', parent=None)
│ Dimensions: ()
│ Data variables:
│ *empty*
│ Attributes:
│ PAL_meta: {"FAC_singlesat": {"dataset": "SW_OPER_MAGA_LR_1B", "model_var...
└── DataTree('SW_OPER_MAGA_LR_1B')
│ Dimensions: (Timestamp: 86400, NEC: 3)
│ Coordinates:
│ * Timestamp (Timestamp) datetime64[ns] 2024-08-24T23:59:59 ... 2024-08-2...
│ Dimensions without coordinates: NEC
│ Data variables:
│ Latitude (Timestamp) float64 -3.933 -3.87 -3.806 ... 65.26 65.2 65.13
│ Longitude (Timestamp) float64 -35.61 -35.61 -35.61 ... 137.5 137.5 137.5
│ Radius (Timestamp) float64 6.847e+06 6.847e+06 ... 6.834e+06 6.834e+06
│ B_NEC (Timestamp, NEC) float64 1.865e+04 -6.146e+03 ... 4.655e+04
│ B_NEC_Model (Timestamp, NEC) float64 1.865e+04 -6.144e+03 ... 4.655e+04
│ Attributes:
│ PAL_meta: {"analysis_window": ["2024-08-24T23:59:59", "2024-08-25T23:59:...
└── DataTree('output')
Dimensions: (Timestamp: 86399)
Coordinates:
* Timestamp (Timestamp) datetime64[ns] 2024-08-24T23:59:59.500000 ... 2024...
Data variables:
FAC (Timestamp) float64 -0.09739 0.001669 ... 0.03192 0.002472
data.swarmpal_fac.quicklook()
(<Figure size 640x480 with 2 Axes>,
array([<Axes: ylabel='d/dt($\\Delta$B)'>,
<Axes: xlabel='Timestamp', ylabel='FAC [uA/m2]'>], dtype=object))
/opt/conda/envs/swarmpal-runner/lib/python3.11/site-packages/IPython/core/events.py:93: UserWarning: Creating legend with loc="best" can be slow with large amounts of data.
func(*args, **kwargs)
/opt/conda/envs/swarmpal-runner/lib/python3.11/site-packages/IPython/core/pylabtools.py:152: UserWarning: Creating legend with loc="best" can be slow with large amounts of data.
fig.canvas.print_figure(bytes_io, **kw)